Looking at enzyme encapsulation and why it matters in bacteria
Bacterial microcompartments are small protein structures that encapsulate various enzymes and substrates. This blog will look at our lab’s recent paper on an enzyme from a bacterial microcompartment, what we learned from it and where to take this research next.
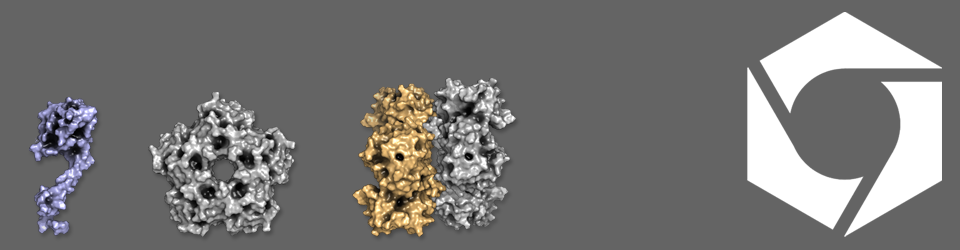
Before discussing why bacterial cells need microcompartments, or even what they are, let’s look at why cells would need to compartmentalise themselves at all. All cell types across all kingdoms of life have some internal structure and capacity for sorting their contents. Cells package their DNA, enzymes for metabolism and chemicals into discreet compartments, with the purpose of making the cell more efficient and protecting it from toxic chemicals produced by the reactions happening inside. Bacterial cells do this via bacterial microcompartments (BMCs), which are protein shells that self-assemble and compartmentalise enzymes and their substrates in a wide range of different bacteria, from gut microbes to bacteria found in the soil and sea.
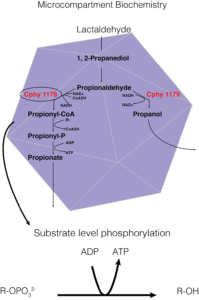
Figure 1. A way of showing the specific biochemistry inside the Clostridium phytofermentans fucose/rhamnose BMC. In this instance, the carbon source is lactaldehyde, Cphy1178 is our aldehyde dehydrogenase enzyme of interest, and the generation of ATP from substrate level phosphorylation is the BMCs main purpose in the cell.
In this instance, the function of BMCs is to protect the cell from toxic intermediates formed from the breakdown of various carbon sources. These carbon sources are used to ‘create’ energy via ATP and the substrate-level phosphorylation pathway, an example of which is given in Figure 1. – the Clostridium phytofermentans fucose/rhamnose microcompartment.
So far, so much chemistry! Much of the appeal for studying BMCs lies in how we may be able to engineer them to use them as cell ‘factories’ to produce biofuels, or to synthesise chemicals that may otherwise be highly toxic to a bacterial cell. However, before we get can properly engineer microcompartments, we need to take a step back and look at what is happening inside them, particularly in terms of their natural chemistry. Our paper looks at the one of the key enzymes in the pathway found in almost all BMCs, the acylating aldehyde dehydrogenase (AldDH). This enzyme takes a highly reactive and toxic aldehyde compound and adds this to an important co-factor coenzyme-A (CoA), which acts to shuttle carbon building blocks between enzymes. This acylated-coA is then used to produce one molecule of ATP in a series of linked reactions that happen in the BMC. We worked with the enzyme from a bacterium called Clostridium phytofermentans, which is related to the Clostridium difficile pathogen that causes hospital acquired infections, but is a harmless soil bacterium that ferments dead plant matter. We show that the enzyme taken from the BMC in this bacteria prefers aldehydes with only a few carbon atoms as its substrate and with the first structure of this type of enzyme with CoA bound we propose a mechanism of action for how this useful enzyme works.
Microcompartment morphology
BMCs are found in various bacterial species, notably pathogenic species of Escherichia coli and Salmonella, where they take advantage of the chemical compounds released from the gut cell wall during their virulent life cycle and use these as a source of energy. Despite encapsulating different metabolic pathways, BMCs in different cells have a similar shape and size, looking like a squashed football about twenty times smaller than the cell. The outer surface of the bacterial microcompartment is pieced together from proteins that share a common basic shape, but tile together as pentagons and hexagons that form the edges and faces of the microcompartment (keeping with the football analogy). The pathway enzymes are held inside the microcompartment, probably around the edge on the inside of the shell proteins (think Kinder Egg), although we aren’t 100% sure of this yet. The enzymes are encapsulated as the microcompartment forms, and are targeted via short address labels known as localisation sequences. Substrates for the enzymes enter via small pores in the shell, which allow products out, but retain the toxic intermediate chemicals produced by the enzymes
Propionaldehdye dehydrogenase – structure with CoA
The aldehyde dehydrogenase (AldDH) in the C. phytofermentans fucose/rhamnose utilisation BMC is on the whole similar to related enzymes in that it uses the NAD+ cofactor to oxidise the aldehyde substrate (in this example the substrate is propionaldehyde) and form an enzyme-linked ester intermediate, producing reduced NADH (Incidentally, the NADH is converted back into NAD+ in the BMC by an alcohol dehydrogenase, meaning that 2 substrate molecules are needed to balance this reaction). However, it differs from other AldDHs in that it uses Coenzyme A to resolve the acyl-enzyme intermediate to produce propionyl-CoA (Figure 1) rather than a glutamic acid general base residue to produce a short-chain fatty acid directly from the enzyme.
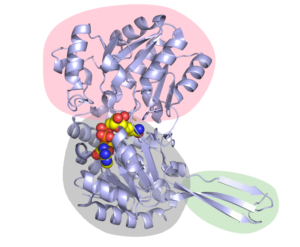
Figure 2. The structure of Cphy1178 AldDH. The pink bubble shows the catalytic binding domain, so the part where the substrate binds. The Rossmann fold nucleotide binding domain is in in grey (and shows NAD+ in the active site as spheres) and the oligomerisation domain is in green (The active form of Cphy1178 is a dimer of dimers).
We found that the CoA molecule binds to the enzyme in the Rossmann-fold nucleotide-binding domain, which is the same pocket that the NAD+ cofactor binds to. Figures 2 & 3, which are taken from the paper, show the configuration of this binding, along with the catalytic binding domain, where the propionaldehyde substrate binds. This in itself is a great achievement, as up until now there is no crystal structure with CoA in the AldDH cofactor-binding site. This information also feeds into our proposed mechanism of action for the enzyme (more on which later).
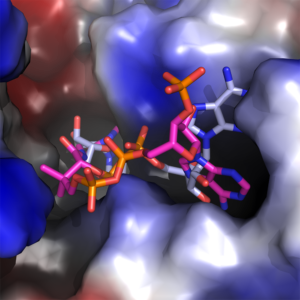
Figure 3. The adenine binding pocket with both NAD+ (blue and grey) and CoA (magenta and orange) configurations shown.
Propionaldehyde dehydrogenase – substrate specificity
Although we had some idea that the substrate for this enzyme is propionaldehyde, the crystal structure of the enzyme shows room for up to a 10 carbon aldehyde, so we decided to quantify the C2-C10 aldehydes’ kcat/KM (which is essentially a test of how efficiently an enzyme reacts with a particular substrate). The assay we used allowed us to follow the formation of NADH from NAD+ by looking at the change in the absorbance of light at a particular wavelength when the enzyme was incubated with different aldehydes and NAD+. From this, we could then calculate the kcat and KM values and give an overall ‘efficiency number’ for each substrate. Experimentally it proved difficult to work with any aldehydes above C6 as they tended to be oily and insoluble in water. Table 1 shows the enzyme efficiency value from each substrate and shows that, as we thought, propionaldehyde is the most suitable substrate for this enzyme.
| Substrate | kcat
(s-1) |
KM
(mM) |
kcat/KM
(s-1 mM-1) |
Ki
(mM) |
| Acetaldehyde | 1.62 ± 0.02 | 5.16 ± 0.10 | 0.31 | n/a |
| Propionaldehyde | 3.45 ± 0.03 | 0.82 ± 0.02 | 4.21 | 17.31 ± 0.46 |
| Butyraldehyde | 3.44 ± 0.7 | 1.74 ± 0.07 | 1.98 | 144.3 ± 30.2 |
| Pentanaldehyde | 5.44 ± 0.10 | 2.2 ± 0.7 | 2.47 | 12.61 ± 0.46 |
| Hexanaldehyde | 5.61 ± 0.12 | 2.90 ± 0.10 | 1.93 | 23.52 ± 1.28 |
Table 1
Propionaldehyde dehydrogenase – mechanism of action
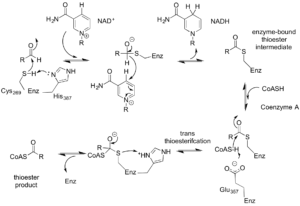
Figure 4. Proposed catalytic mechanism of Cphy1178. The bi-uni-uni-uni-ping-pong mechanism describes a series of electron transfers, converting NAD+ to NADH and Coenzyme A to propionyl-CoA.
From the data generated from the enzyme-CoA crystal structure and the kinetic data, we were able to understand the basis for the proposed mechanism of action for Cphy1178, rather ridiculously named a bi-uni-uni-uni-ping-pong mechanism. We now know that the enzyme resolves the CoA to make an acyl-CoA intermediate, rather than using a glutamic acid general base residue that non-acylating AldDHs do. Figure 4 shows this proposed mechanism and although it seems rather complicated, the main insight is that both NAD+ and CoA cofactors are needed for the full catalytic cycle.
What we’ve learned and next steps
The main take-home message from this paper is that we have the first crystal structure of an acylating aldehyde dehydrogenase with Coenzyme A in the co-factor binding site. We also show kinetic data for this enzyme and show that it can efficiently use aldehydes with up to six-carbon atoms in their fatty chain. In terms of engineering bacterial microcompartments to be a useful synthetic biology tool, it is really important to fully understand the chemistry happening inside of them. In terms of the Clostridium phytofermentans fucose/rhamnose microcompartment, we now know that we could use the propionaldehyde dehydrogenase enzyme inside to turn over longer chain aldehydes, which could be useful in terms of biofuel production. We can also use this information to work out which enzyme we could target to a microcompartment and how it would affect them in terms of overall function.
The next steps for our lab will be to look at formation and shape of microcompartments, their control systems and how much enzyme we can target to them, and if this will increase the reaction efficiency of pathways targeted to BMCs. Hopefully this information will allow us to successfully use BMCs as useful tools for synthetic biology.